How to Determine Which Restriction Enzyme to Use
LA Taq DNA polymerase is. The use of immobilized enzymes finds particular advantages using proteolytic enzymes.

Choosing Proper Restriction Enzymes Based On Defined Criteria For Pcr Download Scientific Diagram
For these reasons where possible select restriction sites that are further apart for cloning.

. Selecting the best NEBuffer to provide reaction conditions that optimize enzyme activity as well as avoid star activity associated with some enzymes is an important consideration. TaKaRa LA Taq DNA Polymerase combines Taq DNA polymerase and a DNA proofreading polymerase with 35 exonuclease activity to enable PCR amplification of very long DNA templates long-range PCR. Other Free Molecular Biology Resources.
I to identify a special enzyme to prove its presence or absence in a distinct specimen like an organism or a tissue and ii to determine the amount of the enzyme in the sample. While for the first the qualitative approach a clear positive or negative result is sufficient the second the. A restriction map is a map of known restriction sites within a sequence of DNARestriction mapping requires the use of restriction enzymesIn molecular biology restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA and sometimes for longer genomic DNAThere are other ways of mapping features on DNA for longer length DNA.
These additional bases provide sufficient DNA for the restriction enzyme to bind the recognition site and cut efficiently. The recognition sequences can also be classified by the number of bases in its recognition site usually between 4 and 8 bases and the number of bases in the sequence will determine how often the site will appear by chance in any given genome eg a 4-base pair. Papain has a variety of industrial applications especially in food industry.
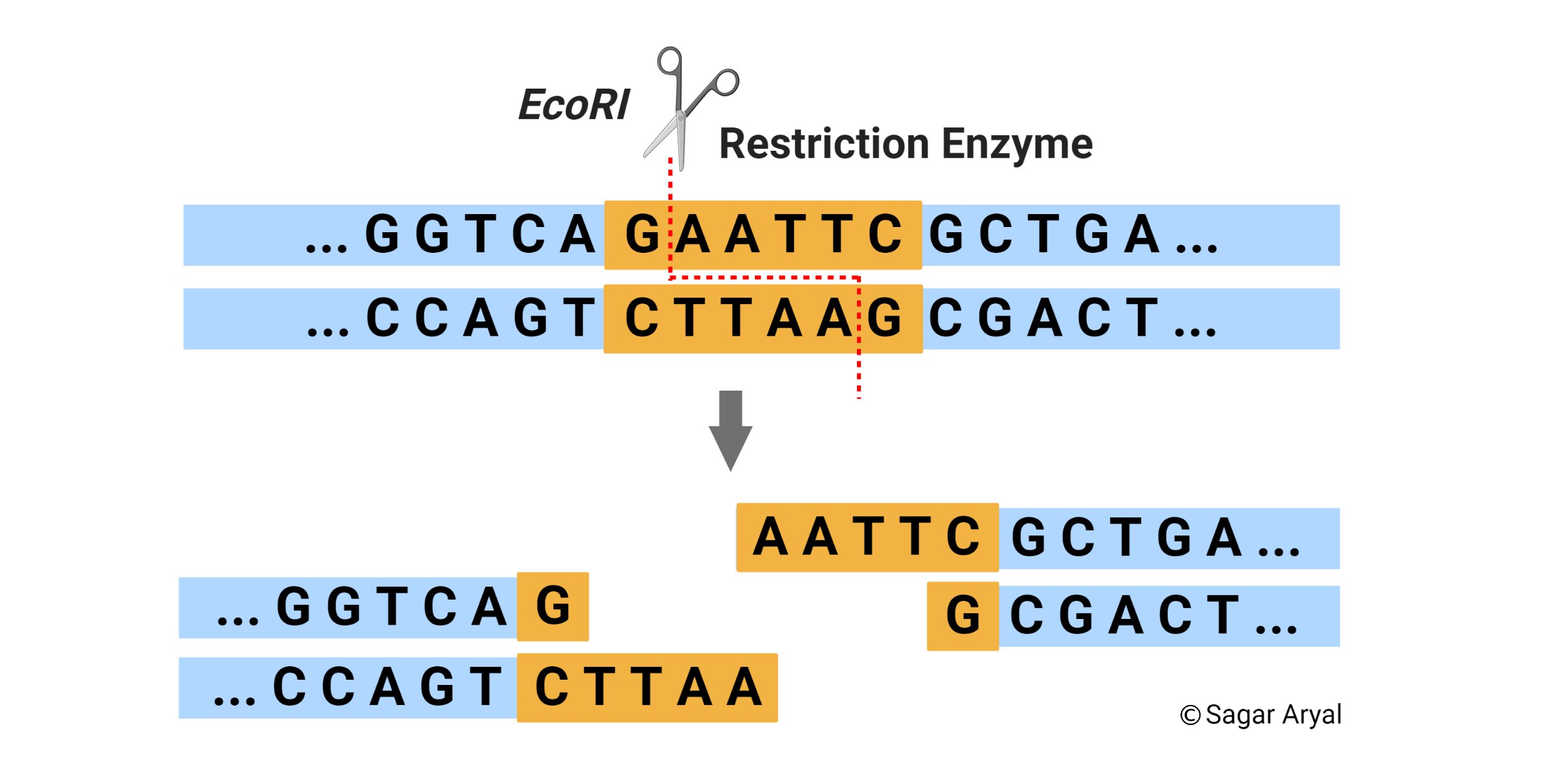
Also does virtual digestion. Digesting a DNA substrate with two restriction endonucleases simultaneously double digestion is a common timesaving procedure. When selecting a restriction sites to add to the primers it is important to determine which sites will be compatible with your selected vector whether directional cloning is desired and most importantly confirm that the recognition sites does not occur within the.
The gene for Sirt6 also called Sirt6 is activated in a wide variety of species meaning there is a large body of research into how Sirt6 uses NAD how it repairs DNA and its role in inflammation and energy metabolism. Maps sites for restriction enzymes aka. Early studies showed that RecA could polymerize on the ssDNA produced by the RecBCD enzyme and could use that ssDNA to produce homologously paired joint molecules 235 239.
Welcome to RestrictionMapper - on line restriction mapping the easy way. And before you set up the reaction determine which enzyme is more effective at cutting close to the end of a DNA fragment and use that enzyme second. Restriction enzymes recognize a specific sequence of nucleotides and produce a double-stranded cut in the DNA.
Subsequently it was demonstrated that Chi stimulated this joint molecule formation in a reaction that optimally. Playing a key part in telomere maintenance DNA repair and gene expression this enzyme is definitely Sirt6 rather than 6 feet under. Enzyme assays are performed to serve two different purposes.
Each enzyme is supplied with its optimal NEBuffer to ensure 100. Restriction endonucleases in DNA sequences. If coadministration cannot be avoided use caution and consider dose reduction of concomitant CYP1A2 substrate drug.
This mixture of enzymes allows for long and accurate LA PCR of targets from a variety of templates including genomic DNA. However if you must use sites in close proximity consider doing a sequential digestion. Concomitant use of vemurafenib with agents with narrow therapeutic windows is not recommended as vemurafenib may alter their concentrations.
Vemurafenib increases levels of tizanidine by affecting hepatic enzyme CYP1A2 metabolism. It is used to. Papain a thiol protease present in the latex of Carica papaya exhibits a broad proteolytic activity and is an enzyme of industrial use and of high research interest.
The type II restriction enzymes cleave foreign DNA into linear fragments that are.

How To Recognize A Recognition Site For A Restriction Enzyme Youtube

How To Recognize A Recognition Site For A Restriction Enzyme Youtube
No comments for "How to Determine Which Restriction Enzyme to Use"
Post a Comment